Convergent evolution between metazoan and choanoflagellate phosphotyrosine signaling
The tyrosine kinase signaling networks of choanoflagellates and metazoans are very distinct, with only a few genes clearly orthologous between the two systems. Yet, both systems seem to have come up with similar or identical mechanisms and architectures. This apparent convergent evolution hints at global constraints within the phosphotyrosine signaling networks. Here are some examples.
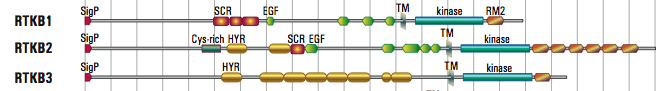
- The 88 receptor TKs (RTKs) in Monosgia lack the immunoglobulin domains found in many human RTKs, but many have related hyalin repeats that are more common in bacteria. Other unique extracellular domains have cysteine-rich motifs that are weakly similar to other regions of metazoan RTKs, suggesting that these are natural structures that can fold outside of the cell and bind to ligands. The RTKB family (some shown below) have a variety of shared and unique extracellular domains.
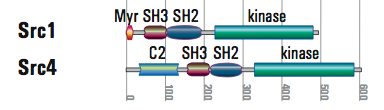
- Src kinases are always tethered to the membrane by myristoylation and the related Tec kinases via a PH lipid-binding domain. One of four Src genes in Monosiga has lost replaced the myristoylation site with a C2 domain, predicted to also bind lipids, but never before seen to anchor a kinase. Same solution, different mechanism.
- Some metazoan TKs have lost catalytic activity, but remain as active genes; independently Monosiga is predicted to have evolved inactive kinases. And both metazoans and Monosiga have independently developed kinase proteins with dual catalytic domains, one of which is inactive. The function of the metazoan kinases (Jaks) is partially understood, and Monosiga may shed light on whether this is a conserved mechanism. Inactive and dual domain phosphatases are also found.

- SH2 domains bind phosphotyrosine and relay the tyrosine kinase signal throughout the cell. SH2 domains couple to many other kinds of domains. Monosiga, covers most of the major classes of metazoan SH2 architectures, but adds many more, mostly with other signaling and adaptor domains, such as SH2, SH3, PDZ, SAM, WW, C1, PH, ankyrin, and EF hand domains. This suggests that choanoflagellates have found new ways to wire the phosphotyrosine signaling circuitry, but that the overall themes are still similar. The few cases of non-signaling domains fused to SH2 proteins found in the initial genome analysis are largely due to gene mis-prediction, with the non-signaling part of the gene separated from the rest by a large intron, indicating that they are separate genes.
Back to the Monosiga kinome page.





